Chapter 5 Overview of Spatial Incidence Models
5.1 Broad Types of Models
Spatial models aim to account for spatial dependencies in the data. These dependencies may not be obvious from the observed data, but spatial data are typically autocorrelated (and/or clustered) such that observations at nearby locations tend to be more similar than observations further away. Spatial models account for this phenomenon by “smoothing” the observed values towards each other. The precise method of smoothing depends on the model.
There are two broad types of spatial models: those that smooth globally, and those that smooth locally. These terms have been given different definitions by different authors. We define them as follows:
- Global spatial smoothing: the same smoothing parameters are applied consistently across the entire region
- Local spatial smoothing: the smoothing parameters can vary from area to area, or from sub-region to sub-region where the sub-regions are either pre-defined (fixed) or estimated by the model.
Using the generic three-stage hierarchical model proposed by Best, Richardson, and Thomson (2005) (see Section 4.2), we now describe specific global and local spatial models that were investigated in the development of the ACA.
5.2 Global Spatial Smoothing Models
5.2.1 Intrinsic CAR and BYM Models
The intrinsic CAR (ICAR) model specifies the following set of conditional distributions for the spatial random effect parameter:
\[ R_i = S_i \\ S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{1}{\sum_j{w_{ij}}}\sum_j{w_{ij}s_j}, \frac{\sigma_s^2}{\sum_j{w_{ij}}}\right) \] where \(w_{ij}\) is the element of a spatial weights matrix \(\bf{W}\) corresponding to row \(i\) and column \(j\) (Besag, York, and Mollié 1991; Besag 1974; Lee 2011; Best, Richardson, and Thomson 2005). The spatial weights matrix determines the spatial proximity between the random effects, and is most commonly defined as a binary, first-order, adjacency matrix, i.e. \[ w_{ij} = \begin{cases} 1 & \mbox{if areas } i \mbox{ and } j \mbox{ are adjacent}\\ 0 & \mbox{otherwise} \\ \end{cases}. \] This model implies that the conditional expectation of \(S_i\) is equal to the mean of the random effects at neighbouring locations.
The \(S_i\) can be regarded as structured spatial random effects. The BYM model (named in honour of the authors Besag, York, and Mollié (1991)) is an extension of the ICAR model which includes an additional parameter for unstructured spatial random effects: \[R_i = S_i + U_i\] where \[U_i \sim \mathcal{N}\left(0, \sigma_U^2\right).\]
Note that there are two identifiability issues with this model which have practical implications.
The first issue is the identifiability of the structured spatial random effects \(S_i\). If an intercept term is included in the model, then these random effects must be constrained to sum to zero, i.e. \(\sum_{i=1}^N{S_i} = 0\). Alternatively, if no intercept is included, then this constraint can be removed. Both parameterisations of the model will provide the same inference. However, note that software like WinBUGS automatically enforces this constraint, so an intercept should be included in this case. Note also that this identifiability issue applies equally to the ICAR model.
The second issue is a likelihood identifiability problem which arises because the two spatial random effects \(S_i\) and \(U_i\) are not uniquely identifiable; only their sum is identifiable (Eberly and Carlin 2000). However, there is a simple remedy which largely resolves this identifiability issue, which can be easily applied post-estimation. This solution requires modifying (or “correcting”) the parameters \(S_i\) and \(U_i\) by an amount that represents the “proportion of excess variation”, variation that cannot be explained by the rest of the model (including any covariates) and therefore ought to be included in the unstructured random effect parameter. Let \(S_i^{(m)}\) and \(U_i^{(m)}\) denote the estimate of the structured and unstructed SREs for the \(m^{\text{th}}\) MCMC iteration respectively. Assuming a sample size of \(M\), the modification is as follows: \[ S_i^{(m)}:=S_i^{(m)} - \psi \cdot U_i^{(m)}, \\ U_i^{(m)}:=U_i^{(m)} + \psi \cdot U_i^{(m)} \] where \(\psi\) is the proportion of excess variation, \[ \psi = \frac{\mathscr{S}(\bm{S})}{\mathscr{S}(\bm{S})+\mathscr{S}(\bm{U})} \\ \mathscr{S}(\bm{S})=sd\left\{\underset{m=1,\ldots,M}{\text{median}} S_i^{(m)} \right\}. \] The following R code illustrates how this simple correction can be implemented. First, we create some dummy data for illustration purposes:
# For illustration, create dummy spatial random effects S and U
M <- 10000
N <- 100 # Assume areas form a 10 by 10 grid
# For 1 iteration
Noise <- rnorm(N, 0, 1.5)
S <- floor((1:N - 1) / 10 - 5) + Noise # Structured, but with some noise
U <- Noise * 2 # Unstructured; noise
# Duplicate results for all M iterations
S <- matrix(S, M, N, byrow = TRUE)
U <- matrix(U, M, N, byrow = TRUE)
# Store the values (for 1 iteration) for plotting
dat <- data.frame(
x = rep(1:10, 10),
y = rep(1:10, each = 10),
val = c(S[1,], U[1,]),
var = rep(c("S", "U"), each = N),
state = "Before"
)For convenience, the computation of the proportion of excess variation \(\psi\) is coded as a user-defined function, which is then applied to each of the random effects. Note the user-defined function assumes \(S\) and \(U\) are two-dimensional arrays of dimension \(M \times N\) where \(N\) is the number of observations (areas).
# User-defined function to compute proportion of excess variation
get.psi <- function(S, U){
sd.S <- sd(apply(S, 2, median))
sd.U <- sd(apply(U, 2, median))
psi <- sd.S / (sd.S + sd.U)
return(psi)
}
# Compute proportion of excess variation
psi <- get.psi(S, U)
# Modify random effects
S <- S - psi * U
U <- U + psi * UViewing the output:
## [1] 0.5325675# Compare the modified random fields S and U
dat <- bind_rows(dat, # Results before modification
data.frame(
x = rep(1:10, 10),
y = rep(1:10, each = 10),
val = c(S[1,], U[1,]),
var = rep(c("S", "U"), each = N),
state = "After"
)
)
ggplot(dat, aes(x = x, y = y, fill = val)) +
geom_raster() +
scale_fill_gradient2(low = "blue", mid = "white", high = "red") +
facet_grid(state ~ var)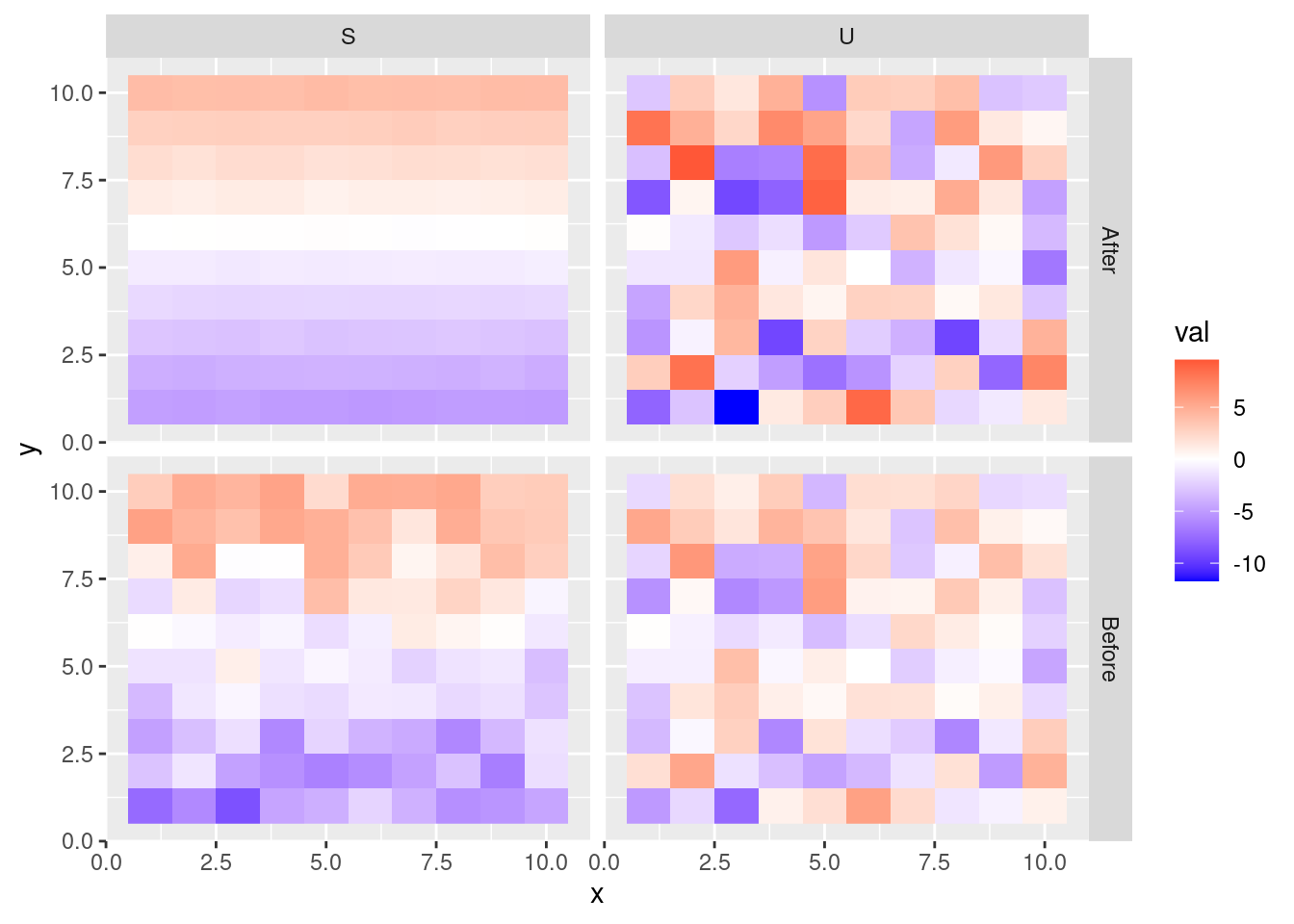
The result is that the structured spatial random effect is now readily identifiable, independently of the unstructured spatial random effect, even though the sum \(S + U\) remains unchanged. The smoothed spatial pattern becomes much clearer after the modification. Assuming the axes represent cardinal directions, the spatial pattern reveals lower values in the South, and larger values in the North. In practice, the modification may not yield such a stark change or seem to improve clarity significantly. However, if there is enough data to identify a structured spatial pattern, this technique will help separate the signal (\(S\)) from the noise (\(U\)).
For further details and an example of a paper which uses this modification so that \(S\) and \(U\) are identifiable separately, see Duncan, White, and Mengersen (2017).
5.2.2 Proper CAR Model
The conditional distributions composing the ICAR prior are proper, but the corresponding joint distribution (not shown here) is improper since the precision matrix is singular. The proper CAR model addresses this by including an additional parameter \(\rho\) which controls the overall level of spatial autocorrelation. The conditional distributions of the proper CAR are: \[S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{\rho}{\sum_j{w_{ij}}}\sum_j{w_{ij}s_j}, \frac{\sigma_s^2}{\sum_j{w_{ij}}}\right)\] where \(\rho\) is subject to the constraint \(-1<\rho<1\), which ensures the precision matrix is positive definite (and therefore the covariance matrix exists), and the joint distribution is proper. An alternative perspective on the parameter \(\rho\) is that it represents the expected proportional “reaction” of \(S_i\) to \(\frac{\sum_j{w_{ij}s_j}}{\sum_j{w_{ij}}}\) (Banerjee, Carlin, and Gelfand 2014).
The proper CAR model is not as popular as the ICAR model because it has several drawbacks: it potentially limits the breadth of the posterior spatial pattern, and for there to be a reasonable amount of spatial association, \(\rho\) likely needs to be close to 1 (Banerjee, Carlin, and Gelfand 2014). In which case, one might as well use the ICAR model, which is more parsimonious (fewer parameters).
5.2.3 Leroux Model
The model proposed by Leroux, Lei, and Breslow (1999) is another variation of the BYM and CAR models discussed above. Like the ICAR model, there is only one spatial random effect parameter for each area,
\[R_i = S_i\]
but the conditional distribution is specified in such a way that it incorporates characteristics of both the structured and unstructured spatial random effects (from the BYM model) into a single parameter (Lee 2011):
\[S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{\rho \sum_j{w_{ij}s_j} + (1-\rho)\mu_0}{\rho \sum_j{w_{ij}}+1-\rho}, \frac{\sigma_s^2}{\rho \sum_j{w_{ij}}+1-\rho}\right)\]
The parameter \(\rho\) plays a similar role to the component weights in a mixture model, except here it weights the type of smoothing: the correlated smoothing of the neighbouring random effects (weighted by \(\rho\)), compared to the uncorrelated smoothing towards a global mean \(\mu_0\) (weighted by \((1-\rho)\)) (Lee and Mitchell 2012). Thus \(S_i\) has a conditional expectation based on a weighted average of both the independent (unstructured) random effects and the spatially structured random effects.
Note that the Leroux model is a generalisation of the ICAR model and the independent model (i.e. a model without any structured spatial random effect). When \(\rho\) is set to 1, the ICAR model is recovered. When \(\rho\) is set to 0, the independent model is recovered. Thus the Leroux model attempts to find balance between these two models by estimating the value of \(\rho\).
It has been demonstrated that the likelihood identifiability issue of the BYM model is not difficult to overcome. However, one advantage of the Leroux model is that it avoids this identifiability issue since there is only one set of random effects, \(S_i\). It also simplifies the difficulty in specifying hyperpriors (in the BYM model, the variance for \(S_i\) is conditional on the neighbouring areas, while the variance for \(U_i\) is marginal) (Riebler et al. 2016)
On a practical note, the formulation for the random effects above includes a global mean, \(\mu_0\). This means an additional overall intercept should not be included in the model, otherwise it will lead to an over-parameterised model and cause identifiability issues. However, the R package CARBayes automatically includes an additional intercept parameter. In this case, the corresponding conditional distributions for the spatial random effects have the global mean term removed (i.e. set \(\mu_0 = 0\)):
\[S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{\rho \sum_j{w_{ij}s_j}}{\rho \sum_j{w_{ij}}+1-\rho}, \frac{\sigma_s^2}{\rho \sum_j{w_{ij}}+1-\rho}\right)\]
Like the BYM model, the random effects in the Leroux model must be constrained to avoid an identifiability issue with the intercept. If the random effects include a global mean, \(\mu_0\), then the random effects must be constrained to sum to \(\beta_0 = \frac{(1-\rho)\mu_0}{\rho \sum_j{w_{ij}} + 1 - \rho}\), else if the model includes the intercept \(\beta_0\) as a separate term in the second stage of the hierarchy, then the random effects must be constrained to sum to zero.
5.2.4 Geostatistical Model
The Geostatistical model proposed by Clements et al. (2006) models the spatial structure as a Gaussian process: \[R_i \sim \mathcal{N}\left(S_i, \sigma^2\right)\] \[S_i=\exp\left(-(\lambda d_{ij})^k\right), \hspace{0.5cm} \lambda > 0\] where \(\lambda\) controls the rate of decay, \(k\) is the degree of spatial smoothing, and \(d_{ij}\) is the distance between points (e.g. centroids of areas) \(i\) and \(j\). The expression for \(S_i\) is the exponential decay function with the addition of the power \(k\). This could be substituted for alternative functions like the disc model (Richardson 1996) (a linear decrease with increasing distance, where two discs of common radius are centred on centroids, and the correlation is proportional to the disc intersection area), or combining two parametric functions to obtain different shapes of decrease, such as the Matern class (Best, Richardson, and Thomson 2005).
The parameters \(\lambda\) and \(k\) are given hyperpriors, usually uniform weakly-informative priors. However, deciding reasonable limits for these priors is often difficult and problem specific.
Perhaps the greatest drawback with this model is the computational complexity. The BYM and CAR models discussed above usually specify the spatial weights matrix in terms of adjacency of areas. This results in the random field \(\bm{S}\) having the Markov property (known as a Markov random field), such that the value of \(S_i\) depends only on a subset of the other areas in the study (i.e. those areas considered neighbours). Conversely, the geostatistical model effectively includes all areas as neighbours of each other area, excluding itself, no matter how far away those neighbours may be geographically. This lack of the Markov property results in a very high computation cost which grows exponentially with the number of areas. (For the ACA investigation of Bayesian spatial methods, it was necessary to impose a “radius of influence” (actually 5 different values of radii, depending on how remote an area was) in order to induce the Markov property and increase the computational efficiency. However, even with this modification, the geostatistical model was prohibitively slow - the slowest model by far).
If a shapefile or similar spatial object is used to define the geographic/administrative boundaries of the areal units, then it is quite easy to compute the distances between the centroids. The following R code shows how to compute the centroids of areas from the “bounding box”, which represents the latitude and longitude of the most extreme points defining a given area in the four cardinal directions. From these centroids, the \(N \times N\) matrix of distances between each pair of centroids is computed.
Note that the shapefile object map is a “spatial polygons data frame”, so the syntax required to interact with this object is a bit different to that required for a regular data frame. For example, to access the polygons information, we use the @ symbol rather than $.
# Import a shapefile; convert to sf
map <- read_sf("Downloaded Data/Shapefile_NZ_Wards.shp")
# Determine the longitude and latitude of centroids
Centroids <- st_centroid(map)
# Compute the distance matrix (Great Circle distance; WGS84 ellipsoid)
d <- st_distance(Centroids, which = "Great Circle")
# Summarise output
dim(d) # d is an N by N matrix## [1] 257 257## Units: [m]
## [,1] [,2] [,3] [,4] [,5]
## [1,] 0.00 80409.78 66730.98 115404.23 122842.91
## [2,] 80409.78 0.00 35410.85 47930.85 43544.11
## [3,] 66730.98 35410.85 0.00 49586.49 64418.67
## [4,] 115404.23 47930.85 49586.49 0.00 29837.28
## [5,] 122842.91 43544.11 64418.67 29837.28 0.00N <- nrow(map)
image(matrix(d, nrow = N)[,N:1], axes = FALSE) # Visualisation of the distances
axis(1, at = seq(0, N, 50)/N, labels = seq(0, N, 50))
axis(2, at = seq(N, 0, -50)/N, labels = seq(0, N, 50))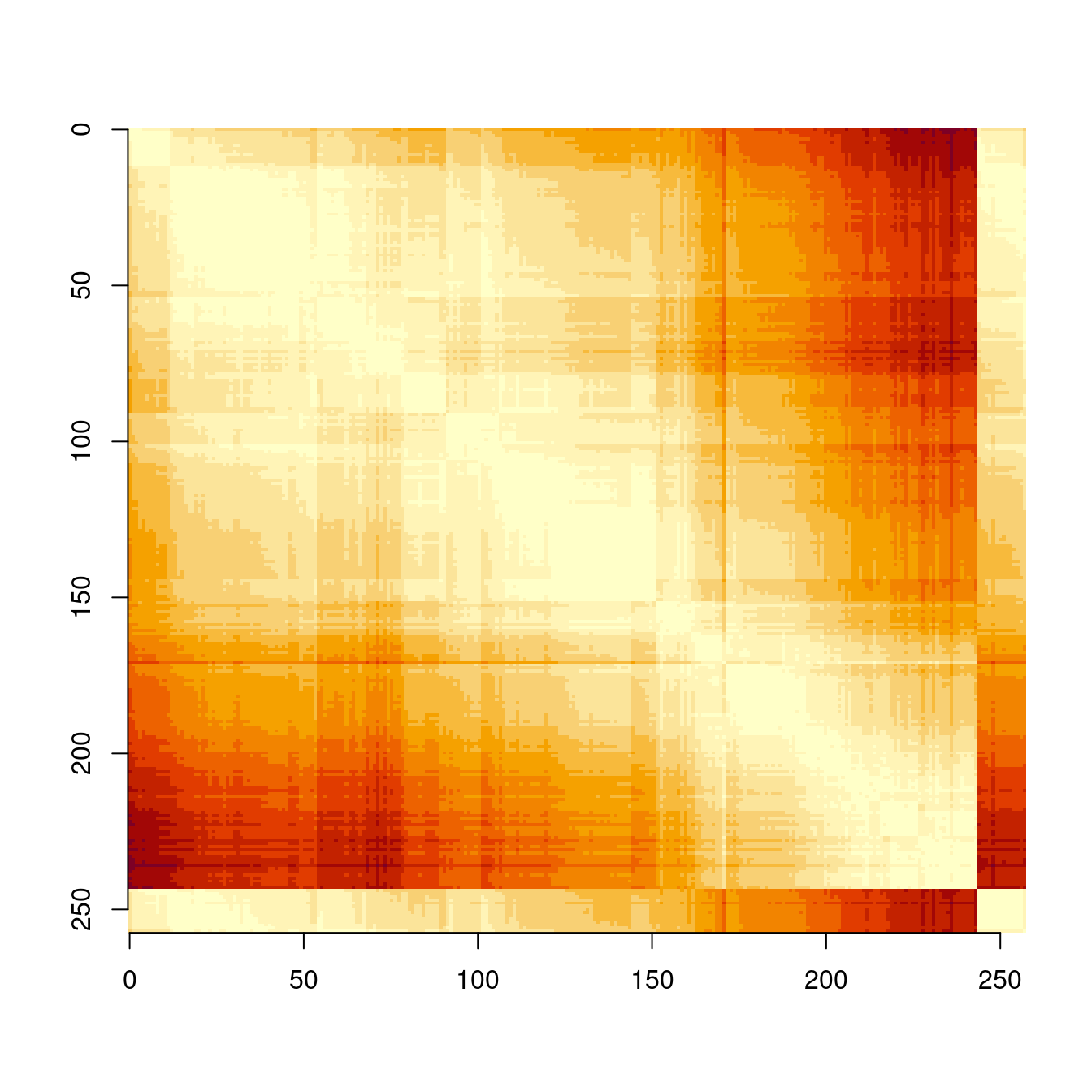
This code may also be useful for other models discussed below, which use distances between centroids as part of the model formulation, such as the CAR dissimilarity model.
5.2.5 Global Spline Models
Global spline models can be divided into two main groups: smoothing splines and P-splines (MacNab 2007). Smoothing splines are penalised splines which have knots on all data points. P-splines allow for a smaller number of knots.
Implementing spline models is not straightforward and requires knowledge on several technically challenging aspects. This includes choosing a smoothing function, selecting the number of degrees of the B-splines, formulating a basis matrix (or penalty matrix from B-splines), selecting the number of knots (and possibly the knot placement). For this reason, we omit further discussion of spline models and direct the reader to Cramb et al. (2017) for further details.
5.3 Local Spatial Smoothing Models
5.3.1 CAR Dissimilarity Model
The CAR dissimilarity model proposed by Lee and Mitchell (2012) is based on the Leroux CAR model, with two modifications:
- the parameter \(\rho\) is fixed at 0.99 to ensure strong global spatial smoothing (while avoiding the impropriety issue of the ICAR model); and
- the spatial weights \(\left\{w_{ij}\right\}\) are estimated rather than fixed which allows the spatial smoothing to be modified locally. (However, the influence between areas is still restricted to immediate, first-order neighbours, and thus the spatial weights matrix \(\bf{W}\) is still sparse).
The similarity between areas \(i\) and \(j\) is determined by including \(q\) non-negative dissimilarity metrics in the model, i.e. \(\bm{z}_{ij} = \left(z_{ij1}, \ldots, z_{ijq}\right)\) where \(\bm{z}_{ijk}=\left|z_{ik}-z_{jk}\right|/\sigma_k\) and \(\sigma_k\) is the standard deviation of \(\left|z_{ik}-z_{jk}\right|\) over all pairs of contiguous areas. The relative impact of the dissimilarity metric(s) is determined by estimating weights in the form of a regression equation. That is, the spatial weights \(\left\{w_{ij}\right\}\) are specified as a function of the dissimilarity metric(s) and their associated unknown regression parameters \(\bm{\alpha}=\left(\alpha_1,\ldots,\alpha_q\right)\). (In the following notation, the dependency on the \(\bm{z}_{ij}\) is suppressed since this is known a priori).
There are two versions of this model, depending on whether the non-zero elements of \(\bf{W}\) are restricted to binary values or not.
The binary formulation is
\[ w_{ij}(\bm{\alpha}) = \begin{cases} 1 & \mbox{if } \exp\left(-\sum_{k=1}^q{z_{ijk}\alpha_k}\right)\geq0.5 \mbox{ and } i\sim j\\ 0 & \mbox{otherwise} \end{cases}, \]
while the non-binary formulation is
\[w_{ij}(\bm{\alpha}) = \exp\left(-\sum_{k=1}^q{z_{ijk}\alpha_k}\right),\]
where in both cases the regression parameters are given a weakly-informative prior
\[\alpha_k \sim \text{Uniform}\left(0, L_k \right) \hspace{0.3cm} \mbox{ for } k=1,\ldots,q \]
and \(L_k\) is a sufficiently large number, fixed a priori. In the CARBayes R package, \(L_k, k=1,\ldots,q\) is fixed at 50 for the non-binary model, and automatically determined based on the distribution of the dissimilarity metric for the binary model. (For the ACA investigation of Bayesian spatial methods, only 1 dissimilarity metric was used, i.e. \(q=1\)).
The conditional distribution for the spatial random effects has exactly the same form as the Leroux model, except \(\rho=0.99\) and \(w_{ij}\) is a function of \(\alpha\), defined by one of the above equations:
\[S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{0.99 \sum_j{w_{ij}(\bm{\alpha})s_j} + 0.01\mu_0}{0.99 \sum_j{w_{ij}}(\bm{\alpha})+0.01}, \frac{\sigma_S^2}{0.99 \sum_j{w_{ij}}(\bm{\alpha})+0.01}\right).\] The two versions of this model (binary and non-binary) can further be distinguished by how the dissimilarity metric(s) \(\bm{z}_{ij}\) are calculated. There are two options:
- Base the dissimilarity metric(s) on measured covariates. This could be social factors, like a socioeconomic index, or physical factors, like physical boundaries (rivers, railway lines, etc.) or distances between centroids. If the aim is to estimate and identify possible boundaries in the risk surface, then the dissimilarity metric(s) should be based on social factors.
- Base the dissimilarity metric on residuals obtained from another model (a two-stage approach). For example, the first stage model could be a BYM or Leroux model, or an independent (i.e. no structured spatial random effect) spatial generalised linear model (GLM). The residuals from this model are used to compute a single dissimilarity metric which is used in the second stage model.
Although this model can have covariates included (i.e. in the second stage of the hierarchy as additive effects, as per Section 4.2), it appears as though there are advantages in excluding them so that the spatial structure is identical in both the risk surface and the random effects surface (Lee and Mitchell 2012).
For the ACA investigation of Bayesian spatial methods, all 4 versions of the CAR dissimilarity model were considered. For the covariate dissimilarity metric, only 1 covariate was used, which was the index of relative socioeconomic advantage and disadvantage (IRSAD) published by the ABS (Australian Bureau of Statistics [ABS] 2013c). For the residual dissimilarity metric model, residuals were obtained from running a Leroux model (where \(\rho\) was estimated, not fixed). Binary and non-binary versions of the weights were used in each case.
5.3.2 Locally Adaptive Model
The locally adaptive model proposed by Lee and Mitchell (2013) is also similar to the Leroux model. The conditional distribution of the spatial random effects is the same as the Leroux model (with \(\mu_0=0\)), where \(\rho\) can be estimated or fixed a priori (Lee and Mitchell (2013) recommend fixing \(rho\) at 0.99). The key difference is the specification of the weights which are estimated rather than fixed.
The spatial weights matrix is initialised as a binary, first-order, adjacency matrix (as defined above for the BYM, Leroux, and CAR models), i.e. \[ w_{ij} = \begin{cases} 1 & \mbox{if areas } i \mbox{ and } j \mbox{ are adjacent}\\ 0 & \mbox{otherwise} \\ \end{cases}. \] but then these weights are updated at each MCMC iteration, while adhering to the restrictions that areas which are not first-order neighbours always have a weight of zero, and the weights of other areas are always binary (either 0 or 1).
Specifically, \(\bf{W}\) is estimated as follows: for adjacent areas \(i\) and \(j\), if the marginal 95% credible intervals of \(S_i\) and \(S_j\) overlap, then set \(w_{ij}=1\), else set \(w_{ij}=0\).
Because only the weights corresponding to first-order neighbours are estimated, this approach should be more computationally feasible than areal wombling (Lu et al. 2007) where all values in \(\bf{W}\) are estimated. However, the computational cost may still be quite high compared to other models where all the weights are fixed, and therefore may necessitate using software like integrated nested Laplace approximation (INLA). However, INLA has its own drawbacks in terms of the statistical inference. Additionally, this model is not “fully” Bayesian since the spatial weights are not treated as individual random quantities within the hierarchical model structure (Lee and Mitchell 2013). For further details, refer to Lee and Mitchell (2013) who implemented this model using INLA.
5.3.3 Localised Autocorrelation Model
The model proposed by Lee and Sarran (2015) assumes an ICAR prior for the spatial random effects with the addition of a piecewise constant intercept. That is, instead of a single overall intercept for the model, the \(N\) areas are partitioned into a maximum of \(G\) clusters, each with their own intercept \((\lambda_1, \ldots, \lambda_G)\). The model is given by:
\[ R_i = S_i + \lambda_{z_i} \\ S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{1}{\sum_j{w_{ij}}}\sum_j{w_{ij}s_j}, \frac{\sigma_s^2}{\sum_j{w_{ij}}}\right) \\ \lambda_g \sim \text{Uniform}\left(\lambda_{g-1}, \lambda_{g+1} \right) \hspace{0.3cm} \mbox{ for } g=1, \ldots, G \\ f\left(Z_i\right) = \frac{\exp\left(-\delta (Z_i-G^*)^2\right)}{\sum_{r=1}^G{\exp\left(-\delta (r-G^*)^2\right)}} \\ \delta \sim \text{Uniform}\left(1, M \right) \]
where \(f(Z_i)\) denotes a shrinkage prior on \(Z_i\) which penalises the values towards the middle intercept value so that the extremes of 1 or \(G\) may be empty. The penalty term \(\delta (Z_i-G^*)^2\) where \(G^*=(G+1)/2\) means that if \(G\) is odd then each data point will be shrunk towards a single intercept \(\lambda_{G^*}\), but if even, there may be two different intercept terms used even if there is a spatially smooth residual structure. Lee and Sarran (2015) thus recommend setting \(G\) to be a small odd number, such as 3 or 5. For the ACA, models using \(G=3\) and \(G=5\) were investigated.
Note that the spatial autocorrelation is accounted for through the ICAR prior on \(\bm{S}\). The clustering is non-spatial in the sense that areas within a given cluster need not be adjacent or geographically close, nor do the clusters need be contiguous. But all areas assigned to the same cluster will have the same intercept. The cluster-specific intercept allows for jumps in the mean surface between adjacent areas if they are in different clusters. The combination of the ICAR prior and cluster-specific intercept yields a form of localised smoothing.
For the ACA investigation of Bayesian spatial methods, the clusters were based on the five categories of Remoteness Areas (major city, inner regional, outer regional, remove, and very remote), published by the Australian Bureau of Statistics [ABS] (2013b)). These remoteness areas are measured using the Accessibility and Remoteness Index of Australia (ARIA+) developed by the University of Adelaide. Note that this measure was originally defined for SA1s. These were aggregated to SA2s by assigning one remoteness category based on population proportions.
5.3.4 Leroux Scale Mixture Model
Congdon (2017) proposed using a scale mixture model within a Leroux prior, as follows:
\[S_i \mid \bm{S}_{\smallsetminus i} \sim \mathcal{N}\left(\frac{\rho \sum_j{\kappa_j w_{ij}s_j} + (1-\rho)\mu_0}{\rho \sum_j{w_{ij}}+1-\rho}, \frac{\sigma_s^2}{\kappa_i \left[ \rho \sum_j{w_{ij}}+1-\rho \right]}\right).\] If \(\rho=0\), this reduces to an unstructured i.i.d. scale mixture Student-t density. Small values of \(\kappa_j (<1)\) will indicate areas differ from their neighbours. The suggested hyperprior for the scale parameter is \[\kappa_i \sim \text{Gam}\left(0.5\nu, 0.5\nu\right)\] where \(\nu\) is a hyperparameter (fixed a priori).
5.3.5 Weighted Sum of Spatial Priors (WSSP) Model
The weighted sum of spatial priors (WSSP) model proposed by Lawson and Clark (2002) can be seen as an extension of the BYM model, where the structured and unstructured spatial random effects \(S_i\) and \(U_i\) are given the same priors as for the BYM model, but the overall spatial random effect is specified as \[R_I = p_i S_i + (1-p_i)Z_i + U_i.\] The \(\{Z_i\}\) random effects model abrupt discontinuities between areas. Lawson and Clark (2002) suggest the following prior for \(\bm{Z}\): \[\pi\left(z_1, \ldots, z_N\right) \propto \frac{1}{\sqrt{\lambda}} \mbox{ exp}\left(-\frac{1}{\lambda}\sum_{i\sim j}\left| Z_i - Z_j \right| \right),\] where \(\lambda\) acts as a constraining term. Note that if \(p_i=1\), the BYM model is recovered, and if \(p_i=0\), then the risk surface is entirely discontinuous.
5.3.6 Other Models
Other models that were initially investigated as part of the ACA investigation of Bayesian spatial methods, but were not implemented include:
- Hidden Potts model (Green and Richardson 2002)
- Spatial partition model Denison and Homes (2001)
- Skew-elliptical areal spatial model (Nathoo and Ghosh 2013)
- Local spline model Goicoa et al. (2012)
Attempts were made to implement the first three models, but without success. The Local spline model was never implemented, largely due to the poor success of the global spline version. Similarly, the proper CAR model was never implemented on theoretical grounds.
5.4 Results from Simulation Study
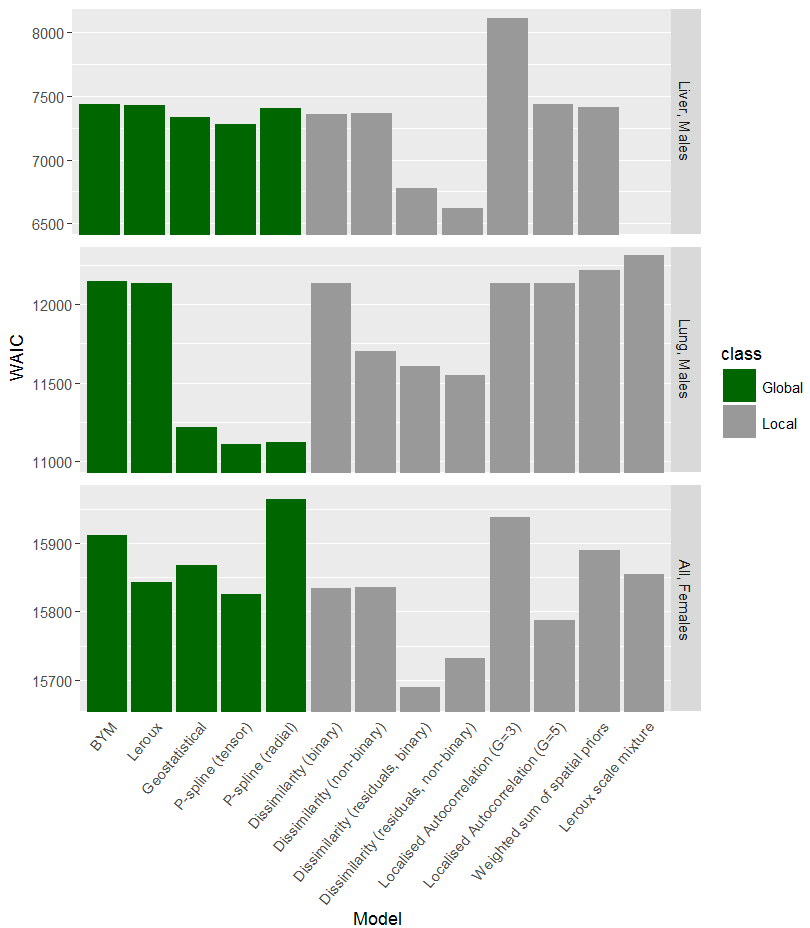
For the geostatistical model, P-spline (tensor) model, both SEIFA dissimilarity models, both locally adaptive models, and (for results pertaining to simulated liver cancer) the Leroux scale mixture model, only limited results are available, such as computation time and model fit criteria. These models were excluded from the final stage of model comparison based on theoretical and practical grounds. The remaining models were ranked across multiple criteria with the aim of identifying the best model to be used for the ACA (Cramb et al. 2017). These criteria are defined and explained in more detail in Chapter 6.
5.4.3 Model Rankings
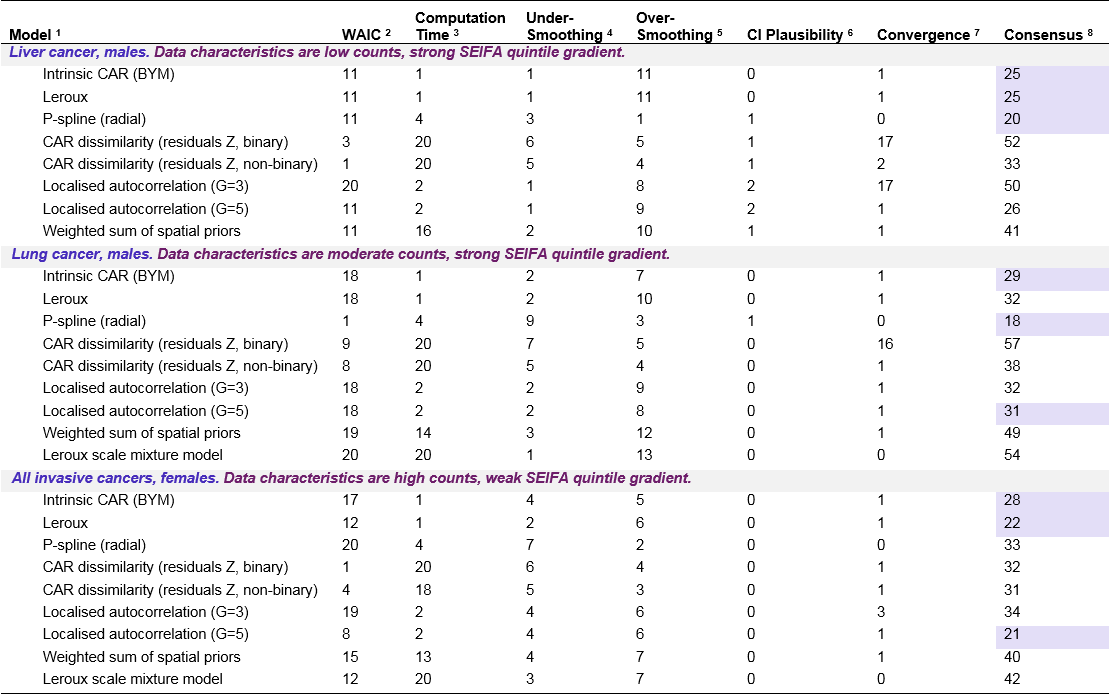
- The Proper CAR model, Hidden Potts model, spatial partition model, local spline model and skew-elliptical model are omitted since they were not run. The Geostatistical model, P-spline (tensor) model, both SEIFA dissimilarity models, both locally adaptive models, and in the case of liver cancer, the Leroux scale mixture model, were excluded on theoretical/practical grounds.
- Rank based on ventiles (20-quantiles) of WAIC (1 = best fit, 20 = worst fit).
- Rank based on ventiles (20-quantiles) of computation time in seconds (1 = fastest, 20 = slowest).
- Rank based on the number of posterior SIRs > 98th percentile of raw SIRs plus number of zero posterior SIRs (since 2nd percentile is zero for all cancers). Smallest rank corresponds to smallest number.
- Rank based on the number of posterior SIRs > 80th percentile of raw SIRs plus number of posterior SIRs < 20th percentile of raw SIRs. Smallest rank corresponds to largest number.
- Ranks: 0 = reasonable; 1 = ok, but some areas are too wide/precise; 2 = bad (either too precise/wide.
- Ranks: 0 = ok (<1% <0.01); 1 = poor (1-<10% <0.01); 2 - 20 = bad: 10%+ <0.01 (Geweke % divided by 5 and rounded)
- The three best models by type of cancer based on the consensus rank are shaded purple.
.png)